Metabolism Tutorial
There is no single model for this tutorial but you will find series of chemical reactions such as the one shown here.
Study the topics in the order shown in the Menu.
Key Concepts
Introduction
Glycolysis is a series of ten reactions that breaks down glucose into smaller molecules that enter the cell's mitochondria. It is within these organelles that these glucose remnants are further decomposed into the waste products carbon dioxide and water. During these later reactions numerous molecules of ATP are produced. ATP provides energy for the vast majority of the cell's activities.
A point often lost when studying each of these reactions -- and trying to keep track of the various atoms -- is the importance of electrons and their accompanying hydrogen nuclei (protons). Essentially, the main theme is that these molecules are stripped of their electrons. These electrons are destined to take part in a series of reactions known as the 'electron transport system'. The electrons are usually removes from their substrates two at a time and accompanied by a matching pair of protons (hydrogen nuclei.)
It is traditional to follow the course of these electrons by watching their hydrogen nuclei partners and this often leads to quite a bit of confusion. Therefore, before beginning these tutorials it will be beneficial to get a firm grip on the various electron/proton arrangements.
Two Types of Hydrogen Ions

The illustration to the right shows how a hydrogen might be removed from an organic molecule. The 'R' stands for the substrate molecule -- whatever it might be -- the : is the pair of electrons that forms a single bond between the substrate and one of its hydrogen atoms. The top picture illustrates this situation.
If the hydrogen is removed -- to 'get at' the electrons bonding it to the substrate -- one of three things might happen:
- The hydrogen nucleus (proton) might be removed leaving both bonding electrons behind. This would result in a negatively charged substrate and the hydrogen would consist solely of it's nucleus (one proton). This is an hydrogen ion (H+).
- The hydrogen might be removed taking only one of the two electrons with it. This would result in both the substrate and the hydrogen being neutral. This is an hydrogen atom (H).
- The hydrogen might be removed taking both bonding electrons with it. This would result in a positively charged substrate and the hydrogen would have two electrons. This is an hydride ion (H-).
Note that the hydrogen ion carries no electrons. These will become part of a 'pool' of hydrogen ions. The hydrogen atom carries one electron and you will find it will be 'transported' around by FAD molecules (see below). The hydride ion carries two electrons and will be 'transported' around by NAD+ (see below).
Oxidation and Reduction
The terms 'oxidation' and 'reduction' are used when there is a transfer of electrons between molecules. The derivation of these terms is awkward and have been explained in three different ways.
- Oxidation is the gain of oxidation and reduction is the loss of oxygen.
- Oxidation is the loss of hydrogen and reduction is the gain of hydrogen.
- Oxidation is the loss of electrons and reduction is the gain of
electrons.
Hint: OIL = Oxidation Is Loss & RIG = Reduction Is Gain.
If there is an electron loser then there has to be an electron gainer -- oxidation and reduction occur together.
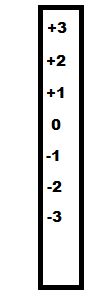
Consider the number scale to the left. If a molecule has a +2 charge then gains an electron its charge will be reduced to +1 on the scale. This is why the term 'reduction' is defined as the gain of an electron -- RIG. Because oxidation and reduction are the opposite of each other, oxidation must be the loss of an electron -- OIL.
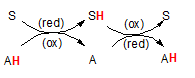
The illustration to the right shows how reduction and oxidation occur together. The 'S' represents a substrate and 'A' represents another molecule we will call an 'agent'. The 'H' is an hydrogen atom attached to these molecules. The hydrogen is colored red to emphasize that this is the atom to keep your eye upon.
The pairs of curved arrows coming together then separating mean the two reactants (S and A) come together then separate. When together the H moves from one to the other. The second pair of curved arrows means they can come together again and transfer the H back again. The first reaction shows the H on the agent (A) meaning it is the source of the H -- and it's electron(s). Because A currently holds the electron(s) it is called a 'reducing agent'. Upon giving these electron(s) to the substrate (S), the substrate has now become reduced but this leaves the agent 'oxidized'. Note the abbreviations (red) and (ox) on the curved arrows. To repeat, the substrate has become reduced while the reducing agent has become oxidized. Follow this same logic in the second reaction. In that case the agent is called the 'oxidizing agent' because it causes the other molecule to become oxidized.
Do not leave this section until you thoroughly understand and can use the terms oxidation, reduction, oxidizing agent and reducing agent.
Transporters / CarriersRemember, an electron is lost from a substrate along with an hydrogen nucleus. Only a hydride ion (H-) and a hydrogen atom (H) carry electrons; the hydride ion carries two while the hydrogen atom carries one. There are two specific transporter that react with these electron-carrying hydrogen nuclei.
FAD
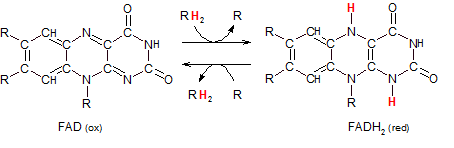
Flavin adenine dinucleotide exists in an oxidized and reduced form. The RH2 entering the top reaction arrow indicates a pair of hydrogen atoms being donated from a substrate (R). Because the substrate is always the 'main player' in reactions it will not be referred to as an 'agent'. It is the other molecule it interacts with that is called an 'agent.'
The intersection of the top two arrows represents the electron-carrying substrate coming into contact with a transporter (FAD). The pair of hydrogen atoms is transferred from one to the other. The 'main player', R, has become oxidized (OIL) so the transporter (FAD) must be the oxidizing agent in this reaction. It becomes reduced to FADH2.
The intersection of the bottom two arrows represents the reaction in reverse. The substrate, R, enters the reaction in it's oxidized (having previously lost electrons) state. The transporter is in it's reduced form FADH2. On contact the hydrogen atoms are transferred from one to the other. The 'main player', R, has gained electrons (RIG) and become reduced while the molecule that 'caused this' -- the reducing agent -- has become oxidized.
NAD+
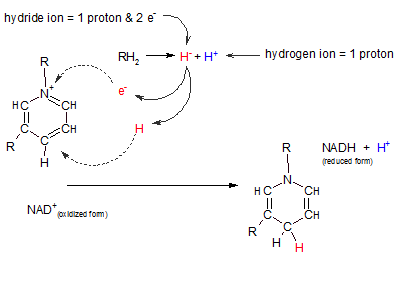
Nicotinamide adenine dinucleotide also exists in two forms: oxidized and reduced. Only the reactive portion of this molecule is illustrated at the right. The "R" region indicates where the remainder of the molecule is attached and is of no importance relative to this discussion.
Notice the following in the illustration to the right:
- The oxidized form is a positively charged molecule (NAD+) owing to the absence of an electron at the position indicated by a plus.
- The reaction at top center (RH2 = H- + H+) represents the loss of two hydrogens from "R". The two electrons are donated together on one hydrogen nucleus (a hydride ion (H-) and the other is simply an hydrogen nucleus(H+).
- When binding with the oxidized form of the transporter (NAD+) the hydride ion loses one of it's two electrons to the positive charge on the transporter (see curved arrow). When this happens the hydride ion becomes a hydrogen atom (H) -- with it's one remaining electron. This atom binds to the transporter where shown by the curved arrow.
- The hydrogen ion (H+) from the substrate remains free and exists in the pool of hydrogen ions .
- Notice the location of the hydrogen atom (red) in the reduced transporter (NADH). Inspect the change in double and single bonds in the transporter.
In this reaction NAD+ acts as the oxidizing agent; it takes electrons from the substrate. In so doing, it becomes reduced and can serve as a reducing agent for some other reaction.
ATP -- the ultimate energy carrier
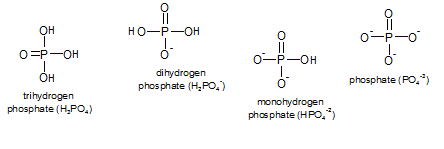
The removal and addition of phosphates are also common types of reactions though they don't necessarily have to occur hand-in-hand like oxidation and reduction. "Phosphates" are a group of small inorganic (not containing carbon) molecules that consist of phosphorus (P) with a double-bonded oxygen and three hydroxyl groups (-OH or -O-). The pH of the environment determines which state(s) will exist and, collectively, they are referred to as inorganic phosphate (Pi). At physiological conditions a mixture of the mono- and dihydrogen phosphates are most common.
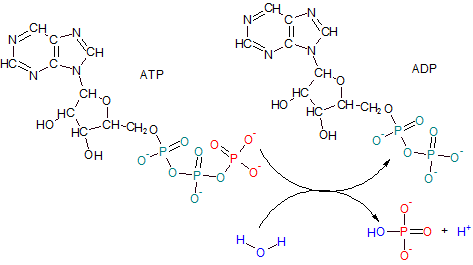
Adenosine Triphosphate (ATP) has a tail of three phosphates--- two teal and one red. ADP results when the terminal phosphate of ATP (red) is removed by the enzyme ATPase leaving only two phosphates behind giving the product the name adenosine diphosphate (ADP). The reaction involves the ionization of water to provide an hydroxyl ion (OH-) (blue) for the released phosphate group and a free hydrogen ion (H+) (blue).
Notice how close together the three double-bonded oxygens are in ATP's phosphate tail. The close proximity of these oxygens creates a dense area of electrons which repel each other. When the terminal phosphate is removed the internal "stress" from this charged region is reduced. The potential energy of the actual bond that was broken to free the phosphate ion is not significantly different from the potential energy of most other bonds. However, the literature often refers to this bond as 'high-energy'. This leaves the incorrect impression that more than a normal amount of energy is released when this bond is broken. Actually, this 'high-energy' reaction refers to the fact that there is significantly less 'overall' energy (including 'stress') remaining in the molecule -- now ADP -- than when it was ATP.
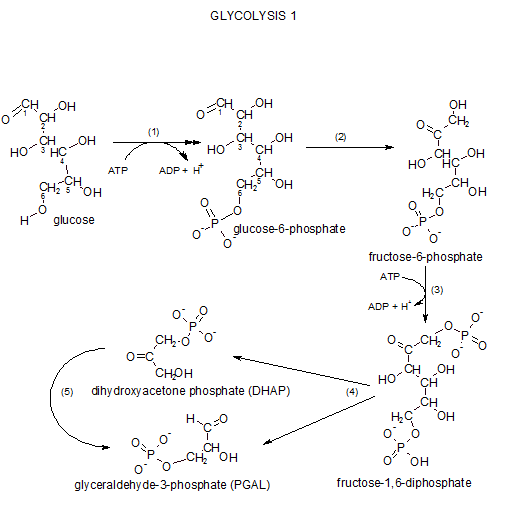
Glycolysis I

The term 'glycolysis' means sugar decomposition or breakdown ('glyco-' is from the Greek for sweet --- glucose used to be named glycose). All reactions occur in the cytoplasm. Glycolysis consists of a sequence of reactions grouped as:
- glycolysis I
- glycolysis II
- intermediate step & Krebs' Cycle
Step 1 ... Phosphorylation
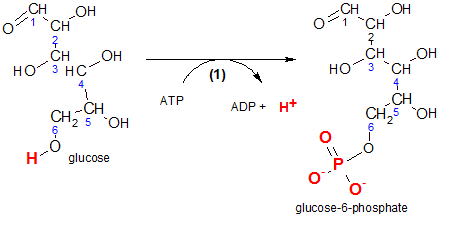
'Phosphorylation' means to add a phosphate group (-PO3-2) to the reactant. Physiologically, this prevents the diffusion of glucose back out of the cell.
Inspect the reactant, glucose, and notice the carbons in its chain are numbered. This numbering notation is important because it gives meaning to some of the chemical names we will encounter. For example, inspection of the product, glucose-6-phosphate, indicates the phosphate group is associated with carbon #6. Verify the two negative charges on that group. These charges are there because these two hydroxyl groups (-OH) have ionized (-O- and H+).
The enzyme responsible for this reaction is hexokinase. 'Hexo' means 'six' because the reactant is a six-carbon molecule and a 'kinase' is an enzyme that catalyzes the phosphorylation of a reactant by ATP. In other words, it transfers a phosphate group from ATP to another molecule...in this case, glucose. It would be helpful at this point if you would quickly review the details of ATP/ADP reactions by clicking here .
The atoms that are involved are shown in red. Comparison of carbon #6 on both the reactant and the product shows that a phosphate group is now located where there once was only a hydrogen. The hydrogen is now a free hydrogen ion shown with ADP.
Step 2 ... Isomerization
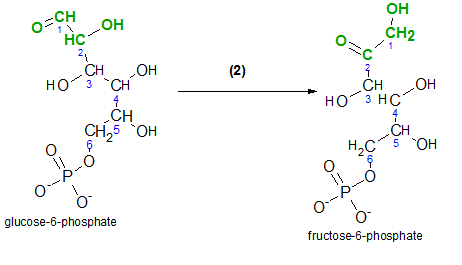
'Isomerization' is a reaction that alters the structural formula of the molecule but does not change the chemical formula. No other molecules take part in this type of reaction.
Begin by comparing the structures of both molecules. The atoms involved in the reaction are shown in green.
- Note that the shape of the carbon chain has been altered; in the reactant, carbon #1 is shown to the left of carbon #2 while it is shown to the right of carbon #2 in the product.
- The reactant is an aldehyde and the product is a ketone.
- An aldehyde is a carbon-chain molecule with a carbonyl group (-CHO). A carbonyl group is a terminal carbon with a double-bonded oxygen and a hydrogen attached. Inspect carbon #1 in the reactant.
- A ketone is a carbon-chain molecule with a double-bonded oxygen on an internal carbon in the chain. Inspect carbon #2 in the product.
- An hydroxyl group (-OH) is on carbon #2 in the reactant and is on carbon #1 in the product.
- There is a single-bonded hydrogen on carbons #1 and #2 in the reactant. In the product there are two single-bonded hydrogens on carbon #1 and none on #2.
If you look closely at the green portions of each molecule you'll notice that the bond between carbons #2 and #3 appears rotated 180 degrees.
The enzyme phosphoglucose isomerase catalyzes this reaction. The ending 'ase' indicates that the molecule is an enzyme and 'isomer' indicates that the molecules involved have the same chemical formulas but different chemical structures. The 'phosphoglucose' part of the enzyme's name identifies the reactant, glucose-6-phosphate.
Step 3 ... Phosphorylation again
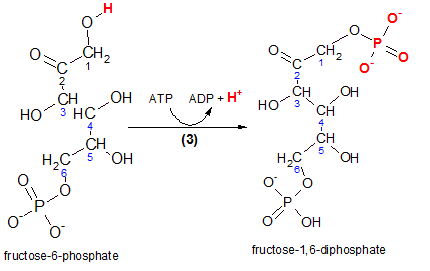
Carefully inspect the reactant, fructose-6-phosphate, in the illustration to the left. The hydrogen (red) is removed and the terminal phosphate group from ATP takes its place as seen in the product molecule. The product is named fructose-1,6-diphosphate and you can verify that the two (di) phosphate groups are associated with carbons #1 and #6. Glance back at the Step 1 reaction and note the similarity.
The enzyme that catalyzes this reaction is phosphofructokinase. Taking the name apart reveals that the reactant, a phosphorylated fructose molecule (phosphofructo), gained another phosphate group from ATP (kinase).
Step 4 ...Cleavage
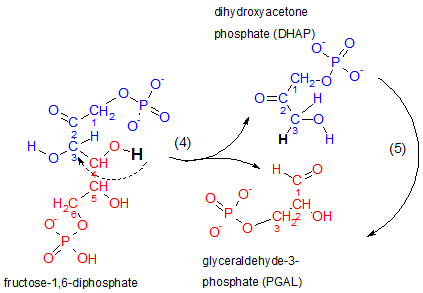
This is the step for which glycolysis is named...'lysis' means 'to separate'. Notice that the reactant breaks between carbons #3 and #4. The top half of the reactant (blue) becomes dihydroxyacetone phosphate, DHAP, and the lower half (red) becomes glyceraldehyde-3-phosphate, PGAL. The hydrogen indicated as black in the reactant loses its association with carbon #4 to become associated with carbon #3 to break the bond between carbons #3 and #4 (see curved dashed arrow).
Inspect the structure of the top product, dihydroxyacetone phosphate. The placement of its double-bonded oxygen classifies it as a ketone. 'Dihydroxy' means 'two hydroxyl (OH-) groups. However, the molecule has only one hydroxyl group on it's #3 carbon...go figure. Acetone (not shown here) is a three-carbon chain with a double-bonded carbon on its #2 carbon and DHAP is similar in this respect thus the inclusion of 'acetone' in this molecule's name. The 'phosphate' part of the name should be obvious.
Inspect the lower product, glyceraldehyde-3-phosphate. It is an aldehyde because the terminal carbon is part of a carbonyl group (-CHO). The double-bonded oxygen of this group is the atom to which the previously mentioned hydrogen atom (black) was attached. The '3' indicates the phosphate is attached to carbon #3. Note the renumbering of carbons in this molecule.
The enzyme responsible for this is aldolase. As always, '-ase' implies the molecule is an enzyme. The 'aldol' part of its name refers to the presence of a double-bonded oxygen with an hydroxyl on an adjacent carbon in the reactant. Verify this by inspecting the atoms attached to carbons #2 and #3 of the reactant.
Step 5...Isomerization again
The enzyme that catalyzes this reaction is triose phosphate isomerase. The curved arrow to the far right in the above illustration indicates that DHAP is internally modified to form PGAL; DHAP and PGAL are isomers as implied by the enzyme's name. 'Triose' means 'a three-carbon sugar'. An easily missed point is that there are now two molecules of PGAL therefore all following reactions will occur twice -- once for each PGAL.
Summary
The six-carbon glucose molecule became two (2) three-carbon molecules (PGAL) that have a phosphate group at one end and a carbonyl group (-CHO) on the other. There was a loss of two (2) ATP molecules to accomplish this. There were two phosphorylation, two isomerization and one cleavage reaction.
Glycolysis II
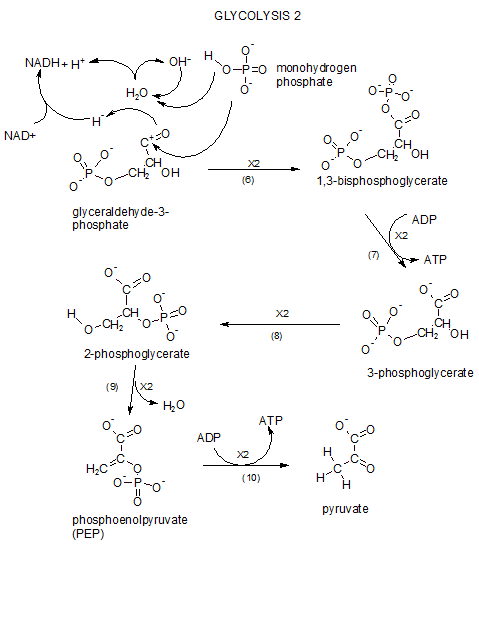
This series of reacions continues the story of how the previously 'lysed' glucose remnants are further broken down. We begin with the two molecules of PGAL which were produced at the end of glycolysis I. Click View Model (above) and print out the glycolysis II reactions to follow during this tutorial. You'll notice a '2X' on each reaction arrow as a reminder that each stem in this sequence occures twice per original glucose molecule.
Step 6...Redox and Phosphorylation
Oxidation/Reduction (redox)
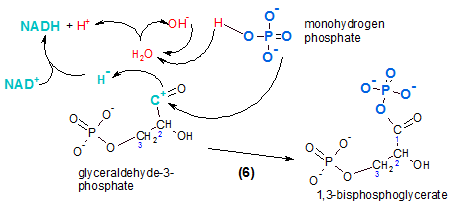
The reactant is PGAL. In the illustration it is shown losing a hydride ion (curved arrow). Several things are going:
- The portion in teal is oxidation/reduction.
- The portion in blue is phosphorylation.
- The portion in red is what happens to a water molecule during the reaction.
Oxidation is the loss of electrons (remember OIL) from the reactant (PGAL). Reduction is the gain of electrons (RIG) by a reducing agent. In this case the reducing agent is the transporter NAD+. Two electrons are lost as a hydride ion. There is a positive charge remaining on the atom where electrons were removed.
The enzyme responsible is PGAL dehydrogenase indicating that PGAL has lost hydrogen. As indicated in a previous tutorial , NAD+/NADH ) a second hydrogen in the form of an hydrogen ion (H+) is often lost from the same molecule. However, in the current reaction, the hydrogen ion is supplied by a water molecule (red). The reduced form of the transporter is NADH.
Phosphorylation
The hydroxyl ion (OH-) (red) from the water molecule is combined with a new hydrogen ion (red) that comes from monohydrogen phosphate (HPO4-2); this generates a new water molecule (note arrows). The remainder of this molecule is the negatively charged phosphate ion (PO4 -3) (blue). This phosphate ion will attach to the positive charge (teal) on the PGAL intermediate (note arrow) forming the product 1,3-bisphosphoglycerate. (The final product has a double-bonded oxygen on #1 with an hydroxyl on an adjacent carbon, #2; this is classified as an aldol.) Its prominent feature is the presence of a phosphate group at each end. Verify that the molecule's name indicates this as well as the carbons to which they are attached. Of future interest is the fact that phosphorylation using an inorganic phosphate adds a fourth oxygen instead of three as when ATP is the source.
Step 7...Dephosphorylation
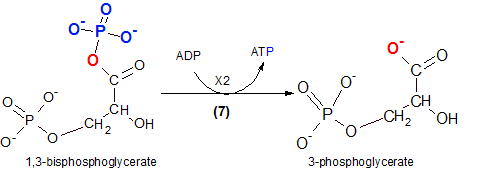
This is our first encounter with the formation of an ATP; it is referred to as 'substrate-level phosphorylation' because the source of ATP's third phosphate group is the reactant (substrate). When the reactant loses the phosphate group (blue) an oxygen (red) remains behind. The phosphate group (-PO3-2) will be picked up by ADP that loses a hydrogen ion (H+) to provide a bonding site. This hydrogen ion becomes part of the hydrogen ion pool. The enzyme that catalyzes this reaction is phosphoglycerate kinase. Remember that a 'kinase' indicates ATP is involved in the reaction.
Step 8...Isomerization
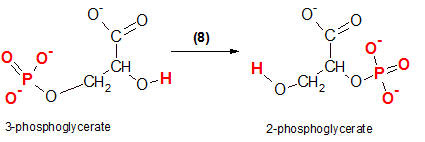
As shown at the left, the reactant, 3-phosphoglycerate, will undergo an internal rearrangement without the lose or gain of any atoms. The product formed will be an 'isomer' of the reactant; they will both have the same molecular formulas but different molecular structures. The product is 2-phosphoglycerate because the #2 carbon now possesses the phosphate group. The red atoms show the before and after locations of the atoms involved.
The enzyme that catalyzes this reaction is phosphoglycerate mutase because it has 'mutated' the reactant.
Step 9...Dehydration
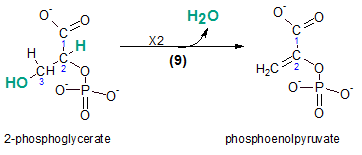
The product of the previous reaction, 2-phosphoglycerate, is dehydrated to form phosphoenolpyruvate (PEP). The atoms lost as a water molecule are shown in teal within the reactant. Compare the bonds between carbons #2 and #3 in the reactant and the product; the single bond has become a double bond.
The enzyme catalyzing this reaction is enolase.
Step 10...Dephosphorylation
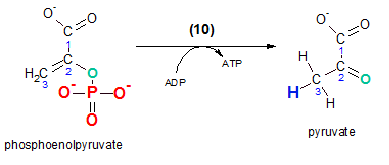
The remaining phosphate will finally be removed leaving behind pyruvate. As in step 7 the acceptor of this phosphate is ADP to form an ATP. Note that the oxygen (teal) that was binding the phosphate group to the reactant is left behind and becomes double-bonded in the product. The third hydrogen (blue) seen on carbon #3 of pyruvate was donated by the ADP molecule providing binding room for its new phosphate group. The enzyme for this reaction is pyruvate kinase ... remember that a kinase involves ATP.
Summary
After each PGAL contributes to the reduction of two (2) molecules of NAD+, each of the two (2) three-carbon molecules receives another phosphate from inorganic phosphates. These are then removed forming two (2) ATP molecules. Shortly thereafter, the remaining phosphate groups are removed to form two (2) more ATP molecules. The end molecules are two (2) three-carbon pyruvate ions.
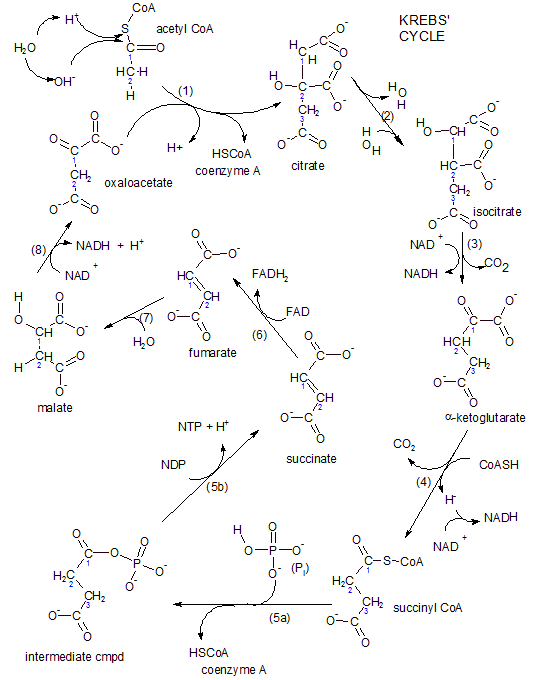
Intermediate Step & Krebs' Cycle
Krebs' cycle, also called the tricarboxylic acid cycle (TCA), consists of a series of carboxylic acids. The first such acid is citric acid that has three carboxyl (-COO-) groups. At physiological conditions these groups exist in this ionic form having lost hydrogen ions to the hydrogen ion pool. Because this series of acids exists as ions they are named, for example, citrate rather than citric acid. Throughout the tutorials we will number the carbon chain without including the carboxyl carbons as part of the numbering scheme.
Breakdown products of all food groups (carbohydrates, fats and proteins) can enter this cycle. However, most must first be converted into a two-carbon molecule of acetyl coenzyme A (acetyl CoA). This conversion is called the 'intermediate step' because it links other pathways to Krebs' Cycle. We begin here with the glucose (a carbohydrate) remnant pyruvate.
The two pyruvates produced at the end of glycolysis cannot cross the inner membrane. There is an transport system embedded within the inner mitochondrial membrane that exchanges one hydroxyl ion from the matrix with one pyruvate ion from the intermembrane space (IMS). Since both pyruvate and hydroxyl ion carry a single negative charge there is no charge (voltage) change across the membrane due to this activity.
Remember that there are two pyruvates produced from each glucose thus, as in glycolysis II, both the intermediate step and the cycle occur twice.
Intermediate Step
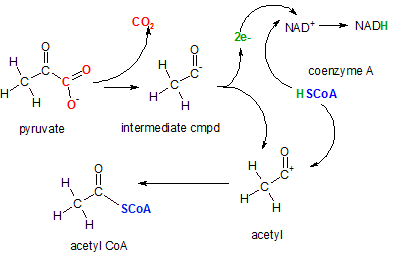
The enzyme pyruvate decarboxylase catalyzes this complex reaction. The first event is the decarboxylation (removal of a carboxyl (-COO-) group) to form carbon dioxide (red). The intermediate compound then donates two electrons (green) to NAD+ to reduce it to NAD- (not shown). Then an hydrogen ion is donated by coenzyme A complete the formation of NADH. The coenzyme A ion (blue) then attaches to the positive charge left on the acetyl ion forming acetyl CoA.
Step 1 ... Synthesis
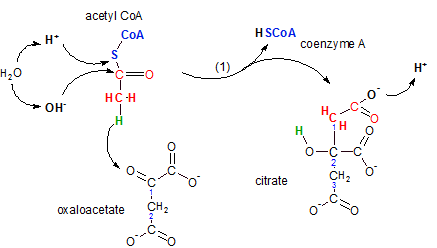
The enzyme citrate synthase catalyzes the joining of the two-carbon acetyl CoA with the four-carbon oxaloacetate to form the six-carbon citrate molecule. During this reaction a molecule of coenzyme A is regenerated -- to participate in another occurrence of the previous step.
Inspect the illustration noting the following:
- Water ionizes
- Acetly CoA is a two-carbon molecule with coenzyme A attached (blue) -- the S (sulfur) is part of the coenzyme
- Oxaloacetate is a two-carbon chain with carboxyl groups (-COO-) attached to carbons #1 and #2 -- it is a four-carbon dicarboxylic acid
- Coenzyme A, when free and not attached to another molecule, has a hydrogen atom attached to it's sulfur.
- Citrate is a three-carbon chain with carboxyl groups attached to each of these three carbons -- it is a six-carbon tricarboxylic acid.
The first event is when the hydrogen (green) on acetyl CoA moves (see curved arrow) to the oxaloacetate's double-bonded oxygen on carbon #1. Verify its new location on the product citrate; it is now part of an hydroxyl group (-OH) on citrate's carbon #2. This move opened a bonding site on acetyl CoA's carbon as well as on carbon #1 of oxaloacetate causing these two bonding sites to combine thus joining the two reactants together.
Confirm that the two carbon atoms (red) in citrate constitute the acetyl part of the new molecule. The synthesis of this six-carbon molecule from a two- and a four-carbon molecule required renumbering of the carbon chain. Notice there are only three carbons in the chain and there are carboxyl groups attached to each of these.
The upper left of the illustration shows the release of the coenzyme occurs when the bond between the sulfur and carbon atoms in the acetyl portion is broken. A water molecule is ionized with it's hydrogen ion bonding to the sulfur and the hydroxyl ion bonding to the carbon. This event releases coenzyme A and also converts the carbon of the acetyl portion into a carboxyl group. Notice that the hydrogen ion is released to join the hydrogen ion pool.
Step 2 ... Isomerization
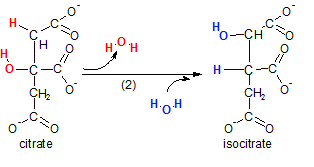
The enzyme aconitase catalyzes this reaction. The reaction actually takes place in two distinct events. Notice that a water molecule (red) is removed (dehydration) then a water (blue) is added back. However, the positions of the hydrogen and hydroxyl groups are now altered. The intermediate compound (not shown) was called 'aconitate'. It is from this transient compound that the enzyme gets it's name. The overall reaction is an isomerization. Verify this by counting the atoms in the reactant (citrate) and the substrate (isocitrate) --- note the type of reaction in the product's name
Step 3 ...Redox & Decarboxylation
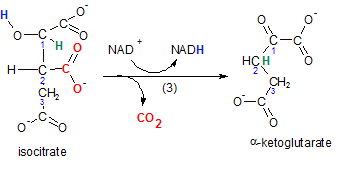
The enzyme for this reaction is isocitrate dehydrogenase because the reactant, isocitrate, loses hydrogen (blue). Isocitrate is oxidized (OIL) while it's oxidizing agent (NAD+) becomes reduced (RIG) to NADH. As always, when the oxidizing agent is NAD+, the reactant loses a pair of electrons as an hydride ion (H-). A second hydrogen (green) is also relocated from carbon #1 to #2. Verify this in the product alpha-ketoglutarate.
The relocation of this hydrogen (green) causes the detachment of a carboxyl group (-COO-) (red). This event is called decarboxylation; carbon dioxide is produced (red).
The product is a three-carbon chain with both a carboxyl and a ketone group associated with carbon #1. Because the ketone is on carbon #1 -- the alpha carbon -- the product is named alpha-ketoglutarate.
step 4 ...Redox & Decarboxylation
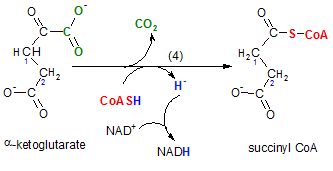
The enzyme for this reaction is alpha-ketoglutarate dehydrogenase. On the surface this reaction looks like a repeat of the previous step. The two reactions are similar in that:
- a NAD+ is reduced to NADH, and
- the reactant loses a carboxyl group to produce carbon dioxide.
Notice the hydrogen (blue) on coenzyme A is released as a hydride ion (blue). As usual, this is picked up by the oxidizing agent NAD+ thus reducing it to NADH. When the carboxyl group (green) is removed the oxidized coenzyme A (red) attaches to that site. The new product, succinyl CoA, is a still a two-carbon chain but it has only one carbonyl group on carbon #2; there are four carbons in all.
Step 5 ... Synthesis
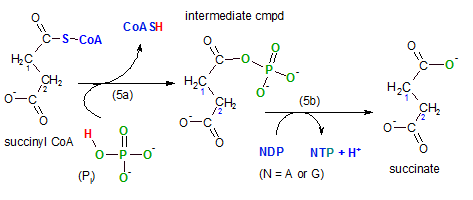
The enzyme for this reaction is succinyl-CoA synthetase; the compound that is being synthesized is either ATP or GTP (guanidine triphosphate) -- depending on the tissue. In the illustration this is shown in reaction (5b) where N (nucleotide) is substituted for either A (ATP) or G (GTP). The hydrogen ion (blue) -- entering the hydrogen ion pool -- is from the NDP to provide a bonding site for the phosphate group (PO3-2) (green) on NTP -- detail not shown. Observe the fourth oxygen (green) from the original inorganic phosphate (Pi) remains on the main substrate. This reaction is a substrate-level phosphorylation reaction as were steps 7 and 10 in glycolysis.
Going back to the first event (5a) in this step we see that the coenzyme A is released from succinyl-CoA. Inorganic phosphate (Pi or HPO4-2) breaks the sulfur-carbon giving it's hydrogen (red) to the coenzyme and it's phosphate ion (green) to the substrate. This forms the intermediate compound shown with the phosphate group where the coenzyme used to be. Notice that the phosphate group now attached has four oxygen atoms but when it is released in (5b) one of these oxygen atoms remains behind with the substrate, succinate.
Comparing the reactant, succinyl-CoA, with the product, succinate, shows there is still a four-carbon molecule with a two-carbon chain having a carbonyl group (-COO-) on each carbon in the chain. The distinguishing part of this reaction is the formation of a high-energy compound, either ATP or GTP.
Step 6 ... Dehydrogenation
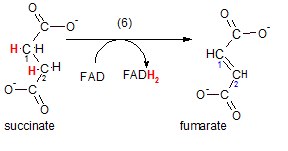
The enzyme is succinic dehydrogenase; it catalyzes the removal of adjacent hydrogen atoms on carbons #1 and #2 of succinate. The product, fumarate, has a double bond between carbon #1 and #2 as a result. The hydrogen atoms take electrons with them so this reaction is also classified as an oxidation (OIL); succinate has been oxidized by the acceptor of the electrons (the oxidizing agent) FAD. As a result, FAD has been reduced to FADH2.
This is our first encounter with this transporter and a good time to review its structure described in Key Concepts. This transporter is unusual in that, instead of floating freely within the mitochondrial matrix, it is attached to the inner surface of the inner mitochondrial membrane. We will encounter this again in the section on the electron transport system.
Step 7 ...Hydration
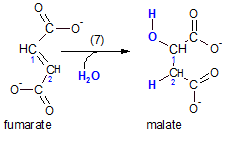
The enzyme fumarase catalyzes this reaction. This is accomplished at the double bond just formed. A water molecule ionizes with the hydrogen ion going to carbon #2 and the hydroxyl ion to carbon #1.
Step 8 ...Dehydrogenation
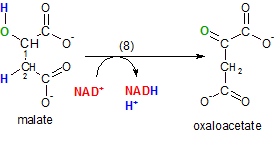
This final Krebs' cycle reaction regenerate the molecule, oxaloacetate, that accepted acetyl CoA into the cycle in step 1. The enzyme malate dehydrogenase catalyzes the removal of the pair of hydrogen atoms (blue) placed on carbons #1 and #2 by water in the previous reaction. Notice that the oxygen (green) of the hydroxyl group is left behind on carbon #1.
NAD+ is the oxidizing agent and, as usual, one of the hydrogen atoms is removed as a hydride ion (not shown) to form NADH and the other hydrogen ion becomes part of the hydrogen ion pool.
Krebs' Cycle Summary
Intermediate Step
This reaction doesn't belong to either glycolysis or Krebs' cycle; it is an 'intermediate' step. Glycolysis ends with the production of two three-carbon pyruvate molecules. These cannot enter Krebs' cycle so they are decarboxylated to two-carbon acetyl-CoA molecules. Two molecules of NADH are also produced. Each acetyl-CoA molecule can now enter the Krebs' cycle.
Krebs' Cycle
This is a series of eight reactions with the last product, oxaloacetate, being the same as the first reactant, thus the term 'cycle'.
- This step combines a two-carbon with a four-carbon molecule to form the six-carbon citrate molecule and releasing the coenzyme.
- Isomerization rearranges the atoms within the molecule changing it's structure.
- Decarboxylation releases carbon dioxide. The reactant is also oxidized and NADH is produced.
- Decarboxylation releases a second carbon dioxide. The reactant is also oxidized again and NADH is produced. Coenzyme A is attached to the substrate.
- A phosphate group replaces the coenzyme then it leaves the substrate becoming attached to a nucleotide diphosphate, either ADP or GDP. The triphosphate form is produced.
- The substrate is oxidized producing FADH2.
- The substrate is hydrated.
- The substrate is oxidized producing NADH and regenerating the oxaloacetate molecule.
Summary of the Summary
In essence, it's as if each two-carbon acetyl-CoA entering the cycle from the intermediate step leaves the cycle as two (2) carbon dioxide molecules. The initial uptake substrate, oxaloacetate, is regenerated. Three (3) molecules of NADH are produced. One (1) molecule of FADH2 is produced. One (1) high-energy nucleotide molecule (NTP) is produced by substrate-level phosphorylation. Coenzyme A plays several roles in these reactions.
Because the original glucose molecule produces two molecules for the intermediate step the above counts must be doubled. Combining those tallies with the ones from glycolysis we get the following products:
- CO2 -- none from glycolysis, two from the intermediate step and four from the cycle totals 6.
- ATP -- two from glycolysis, none from the intermediate step and two (NTP) -- count as ATP -- from the cycle totals 4.
- NADH -- two from glycolysis, two from the intermediate step and six from the cycle totals 10.
- FADH2 -- two from the cycle totals 2.
Electron Transport System & Oxidative Phosphorylation
The details of the molecular complexes involved in this set of reactions have recently been illuminated in great detail. However, the mechanisms of the hydrogen ion 'pumps' associated with complexes I, II and IV are still being investigated. Keep in mind that, at this point, the literature still contains many versions of how this system works and the number of ATP molecules resulting per breakdown of one glucose molecule. This summary will only consider the system as presented here.
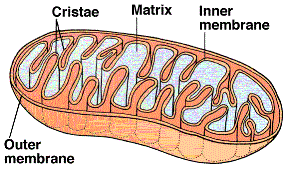
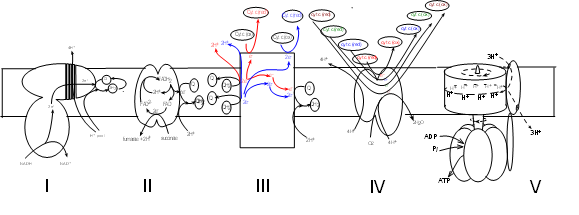
The final phase occurs within the mitochondria. The cut-away illustration of a mitochondrion shows a large inner membrane with folds called cristae. The center is a fluid-filled matrix that contains the soluble enzymes of Krebs' cycle. There is a narrow space between the inner and outer membranes (not labeled) called the intermembrane space (IMS).
The schematic illustrations shows five large protein complexes that are embedded within the inner mitochondrial membrane. Current research suggests several ways they might be grouped but there is no consensus. Many copies of each complex likely diffuse slowly within the membrane. Also within the inner membrane are small hydrophobic molecules of coenzyme Q that diffuse more rapidly. Water-soluble molecules of cytochrome c circulate within the intermembrane space (represented at the top of the illustration). The mitochondrial matrix would be at the bottom of the illustration where the Roman numerals are shown.
If considered as a set, the electron transport system (ETS) consists of four (I-IV) complexes plus many molecules of coenzyme Q and cytochrome c. Complex V takes part in 'oxidative phosphorylation' -- the final step. The details of this illustration are too small to read at this resolution but each will be fully drawn out in the following tutorials.
Complex I
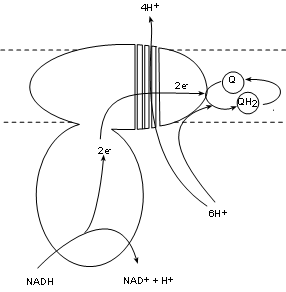
Complex I, also called NADH dehydrogenase, has a binding site for NADH that hangs into the matrix. Two electrons are transferred into the complex while the hydrogen ion and the oxidized NAD+ are released into the matrix. The electron pair is passed between several molecules within the complex to the complex surface buried within the inner membrane. There is a bonding site there for coenzyme Q.
Existing within the hydrophobic interior of the inner membrane are small mobile molecules of coenzyme Q. As common with electron transporters these have 'tag along' hydrogen nuclei (H+). These hydrogen ions are picked up directly from the matrix pool and join the electron pair to fully reduce coenzyme Q to QH2. As this is occurring there is a conformational change -- possibly forming four channels -- that 'pumps' four (4) hydrogen ions from the matrix pool into the intermembrane space.
The key points are:
- one (1) hydrogen ion and NAD+ to the matrix.
- one (1) pair of electrons that the complex transfers to coenzyme Q forming Q-2 (not shown).
- Two (2) hydrogen ions from the matrix combine with Q-2 to form QH2.
- Four (4) hydrogen ions are 'pumped' from the matrix into the intermembrane space.
Complex II
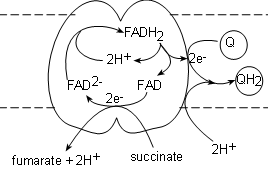
Complex II, also called succinate dehydrogenase , contains the actual enzyme we saw in step 6 of Krebs' cycle. It's binding site for succinate is on the matrix side of the membrane and the transporter (FAD) is actually part of the complex itself. The electrons from the succinate reduce FAD to FAD-2 while the hydrogen ions and fumarate are released into the matrix.
A FADH2, from within the complex, is oxidized producing a pair of hydrogen ions to complete the reduction of the newly-formed FAD-2 thus regenerating itself. The FAD will accept the next pair of electrons that are received. The electron pair is transferred to a coenzyme Q attached to the surface of the complex to form Q-2. Finally, a pair of hydrogen ions from the pool in the matrix completes the reduction of the coenzyme to QH2.
The key points are: One succinate molecule is reduced to donate:
- One (1) pair of hydrogen ions and fumarate to the matrix.
- One (1) pair of electrons to the FAD within the complex that are eventually donated to one (1) coenzyme Q forming Q-2 (not shown)
- One (1) pair of hydrogen ions from the matrix combines with the reduced Q-2 to form one (1) QH2.
Complex III
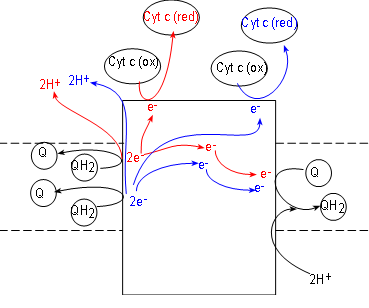
Initial reactants for this enzyme complex are coenzyme QH2 molecules (shown at the left side of the complex). These have been accumulating in the hydrophobic interior of the inner membrane due to the activities of complexes I and II.
Complex III, also called coenzyme Q - cytochrome c reductase. simultaneously reduces both coenzyme Q (illustrated at the right of the complex) and cytochrome c (illustrated at the top of the complex). Cytochrome c molecules are numerous and located within the IMS. Like complexes I and II, complex III also reduces coenzyme Q to QH2 (at the right of the complex).
To understand what occurs we must follow one QH2 at a time. Inspect the top QH2 at the left of the complex. Notice it contacts the surface of the complex to release a pair of electrons (red) to the complex and the pair of hydrogen ions (red) into the IMS.
Following the electrons (red arrows) notice that:
- One electron is transferred to the complex surface where it is picked up by an oxidized cytochrome c (these receive only one electron). Being reduced (red), this cytochrome detaches from the complex surface and returns to the IMS,
- The second electron (red) is transferred to the embedded part of the surface of the complex. Here it will combine with a coenzyme Q attached there. This Q, with a single electron (not shown), will remain attached to await the arrival of a second electron.
Now follow the products (blue) of the second QH2. A second pair of hydrogen ions is added to the intermembrane space. Another electron (blue) reduces a second cytochrome c. The second electron (blue) is transferred to the half-way reduced coenzyme Q plus a pair of hydrogen ions from the matrix completes the formation of a molecule of QH2. It is released from the surface of complex III to return to the hydrophobic interior of the membrane -- and complexes I and II.
The hydrogen ion 'pump' of this complex is indirect. In complexes I (and IV) hydrogen ions are physically moved from the matrix to the space. In this case two pairs of hydrogen ions are moved out of the interior of the inner membrane while one pair is moved from the matrix into the interior of that membrane. *This is equivalent to moving a pair from the matrix to the space. Complex II is credited with pumping one pair of hydrogen ions into the intermembrane space instead of two.
The key points are: For each pair of QH2 oxidized:
- One (1) molecule of coenzyme Q is reduced.
- One (1) pair of hydrogen ions is removed from the matrix to help reduce Q-2.
- One (1) pair of hydrogen ions are moved from the matrix to the intermembrane space.*
- One (1) pair of cytochrome c transporters are reduced (one electron each).
Complex IV
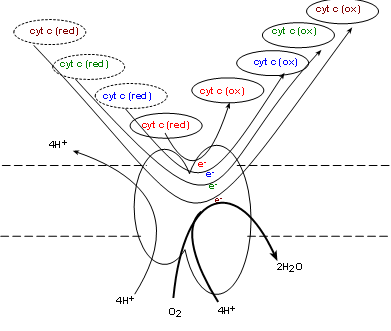 Complex IV, also called cytochrome c oxidase (COX) . will oxidize the reduced coenzyme c molecules produced
by complex III returning the oxidized forms to the intermembrane space -- and to complex III. Simultaneously, one molecule of
molecular oxygen (O2) from the matrix is taken into the complex. Current research indicates
four electrons, donated by four cytochrome c molecules, break the double-bond between the oxygen atoms
immediately forming two oxygen ions (O-2). The mechanism is unclear as to how four hydrogen
ions from the matrix are simultaneously
pumped into the intermembrane space. Each oxygen ion receives a pair of hydrogen ions from the matrix to
form water.
Oxygen is known as 'the final electron acceptor' reducing molecular oxygen to water.
Complex IV, also called cytochrome c oxidase (COX) . will oxidize the reduced coenzyme c molecules produced
by complex III returning the oxidized forms to the intermembrane space -- and to complex III. Simultaneously, one molecule of
molecular oxygen (O2) from the matrix is taken into the complex. Current research indicates
four electrons, donated by four cytochrome c molecules, break the double-bond between the oxygen atoms
immediately forming two oxygen ions (O-2). The mechanism is unclear as to how four hydrogen
ions from the matrix are simultaneously
pumped into the intermembrane space. Each oxygen ion receives a pair of hydrogen ions from the matrix to
form water.
Oxygen is known as 'the final electron acceptor' reducing molecular oxygen to water.
The key points are: For every four (4) cytochrome c molecules that are oxidized the following occurs:
- Four (4) cytochrome c molecules are reduced
- Four (4) hydrogen ions are pumped from the matrix into the intermembrane space.
- One (1) molecular oxygen is reduced to water.
- Four (4) hydrogen ions are removed from the matrix to combine with oxygen.
Shuttles
There are two (2) NADH molecules produced in the cytoplasm during glycolysis. They cannot cross the inner membrane. In order to utilize their potential to donate electrons they use one of two shuttle systems. These shuttles are self replenishing so as not to run out of participating molecules.
Malate-Aspartate Shuttle
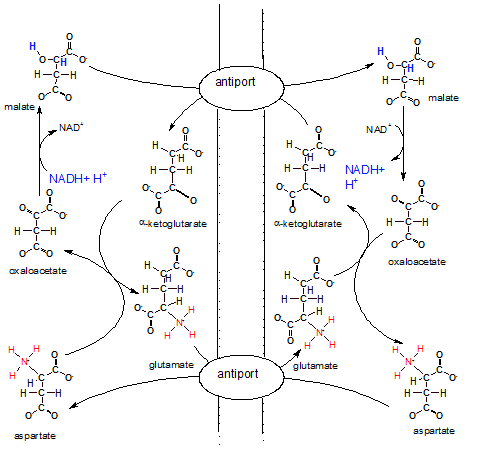
The illustration shows the inner membrane placed vertically with the IMS on it's left and the matrix on it's right. There are two antiports (exchangers) embedded in the inner mitochondrial membrane that swap molecules across the membrane.
The overall big picture of all these reactions looks like two circles; a smaller one inside a larger one. The arrows of the larger outer circle go clockwise while those of the inner one go counter-clockwise.
Notice NADH + H+ in the IMS reducing oxaloacetate to malate. Malate now is the bearer of the electrons and hydrogens with which we are concerned. To move these electrons and hydrogens -- within malate --to the matrix there must be an alpha-ketoglutarate bound to the antiport molecule. Only when both molecules --malate and alphaketoglutarate -- are attached will the antiport simultaneously move them to opposite sides of the membrane. Once in the matrix the enzyme for step 8 of Krebs' cycle converts the malate to oxaloacetate releasing NADH and H+. Mission accomplished. Now NADH can donate it's electrons to complex I of the ETS.
The matrix loses a molecule of alpha-ketoglutarate, an important substrate in Krebs' cycle, during the above event. To compensate for this loss a new molecule is formed when oxaloacetate (formerly malate) reacts with glutamate that has been transferred in from the IMS. During this reaction aspartate is formed. An antiport for glutamate and aspartate insures a continuing supply of glutamate for future matrix reactions.
Inspection of the IMS side of the membrane shows that alpha-ketoglutarate is converted to glutamate to insure it's continuing supply. Since it is moved to the matrix by an antiport there must also be a continuing supply of its 'partner' aspartate. Aspartate is produced from oxaloacetate as mentioned earlier. Also notice that once in the IMS the aspartate is converted to oxaloacetate to continue the outer-circle reactions. A lot of work to move regenerate a single NADH on the matrix side of the membrane.
Glycerol Phosphate Shuttle
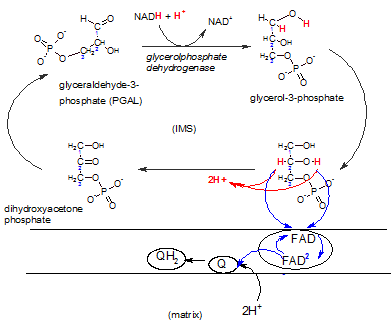
This shuttle system is primarily found in brown fat and infants. A NADH produced in the cytoplasm during glycolysis can react with DHAP reducing it to glycerol-3-phosphate that now carries the electrons and hydrogens. The enzyme complex embedded in the inner membrane (lower right) contains the enzyme FAD-dependent glyceolphosphate dehydrogenase. There is a binding site for glycerol-3-phosphate on it's IMS side. The enzyme oxidizes this reactant while reducing FAD that is an integral part of the complex. Notice that FAD accepts only the electron pair (blue) ; the hydrogen ion pair (red) is set free in the IMS. The FAD-2 then reduces a coenzyme Q within the hydrophobic inner membrane ; this picks up a pair of hydrogen ions from the matrix to form QH2 -- now available to react with complex III of the ETS.
The difference between using this shuttle as opposed to the previous shuttle is that the previous one releases the electron pair to complex I -- early in the ETS -- while this shuttle releases the electrons to complex III.
Shuttle Summary
The point of knowing which shuttle is used to capture the electron-transferring power of cytoplasmic NADH molecules comes into play when trying to determine the number of ATP molecules formed as a result of events occurring in the ETS. This will be discussed after the following section. The key point is that if the malate-aspartate shuttle is used there is a NADH regenerated from a NADH. If the glycerol phosphate shuttle is used a QH2 is produced. NAD enters the ETS at complex I so that it's electron pair will pass through all three proton-pumping complexes. However, QH2 gives it's electron pair to complex III bypassing the proton-pumping power of complex I.
Oxidative Phosphorylation
This group of reactions utilizes the hydrogen ion gradient across the inner membrane to produce ATP. The term 'oxidative' refers to the fact that the hydrogen ions were concentrated on the IMS side of the membrane due to the oxidation of the reduced transporters previously formed. 'Phosphorylation' refers to the addition of a phosphate group to ADP forming ATP.
Complex V
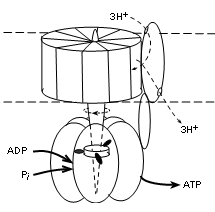
The illustration shows that this complex has two major parts; that embedded within the membrane (between dotted lines) and that suspended into the matrix (lower part of illustration). The embedded part has a central shaft that extends into the matrix part. This complex is actually a motor -- it moves! The embedded part rotates -- staff included. The outer portion of the matrix part is stationary but the shaft within it does move. The 'fuel' for the rotation is the diffusion of hydrogen ions, along their electrochemical gradient (proton motive force), from the IMS (top of illustration) to the matrix.
Current literature reports ten (10) proteins encircling the shaft within the membrane in human mitochondria. When there are ten, research suggests that a set of three (3) hydrogen ions, diffusing through a separate protein 'channel' will turn the membrane portion 120 degrees. It appears that the number of proteins encircling this part of the shaft determines the number of hydrogen ions required to produce this rotation.
There are three (3) sets of protein pairs (alpha and beta, not labeled) that fit side-by-side surrounding the shaft in the matrix part. The rotation of the shaft causes shape changes of the binding sites in these pairs.
The shaft between the three unit pairs has three 'spikes' arranged so that one contacts each pair simultaneously (see illustration). With each 120 degree rotation the spikes rotate to contact the adjacent alpha/beta pair. Another 120 degree rotation moves them to the next pair, etc. When spike #1 contacts a unit physical conformation of the pair changes to allow an ADP and inorganic phosphate to enter. When spike #2 contacts this pair the bonding sites changes again to bring the reactants into contact so ATP can be synthesized. Contact with spike #3 separates the pair to release ATP and the return of spike #1 alters the conformation to accept the next pair of reactants.
Each alpha/beta pair is 'one-step-behind' the one before it thus there is an ATP released with each 120 degree turn of the shaft. The stoichiometry (i.e., relation between the amount of two things) between the diffusion of three (3) hydrogen ions is the production of one (1) ATP -- 3H+:1ATP
Translocators
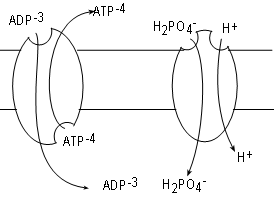
ADP is formed in the cytoplasm and has too great a negative charge to pass through the inner mitochondrial membrane. Likewise, ATP formed within the matrix can not pass through either for the same reason. Many copies of a transporter called adenosine nucleotide translocase (ANT), embedded in the inner membrane, serve the exchange one molecule of ADP for one molecule of ATP.
The mechanism for the exchange involves conformational changes. At any time a bonding site exists on one side or the other but not both simultaneously as implied by the illustration. A site on the matrix side will bind ATP. Once bound the molecule changes to evert the ATP to the IMS. After that a new site for ADP forms on the IMS side. When an ADP binds the eversion takes place in reverse. Take note that the exchange creates a change in the charge (voltage) difference across the membrane.
Inorganic phosphate is also in the cytoplasm but cannot pass through the membrane due to it's charge. Multiple copies of the translocator phosphate translocase, embedded in the membrane, relocate one inorganic phosphate plus a hydrogen ion from the IMS into the matrix. In this case both substrates move in the same direction. This is termed 'symport'; both substrates must be bound to activate the 'pump'. The driving force is the hydrogen ion gradient favoring transport from the IMS to the matrix. Current literature favors counting the transfer of this hydrogen ion into the matrix along with the three moving through complex V when tabulating the number of ATPs formed.
ATP Synthesis
Synthesis of ATP required the movement of hydrogen ions along a gradient moving them from the IMS to the matrix. This gradient is produced by the pumping of hydrogen ions into the IMS as described above. We now believe it requires the movement of four (4) hydrogen ions -- three through complex V plus one via the phosphate/hydrogen ion symport -- back into the matrix to synthesize one (1) ATP. By tabulating the number of hydrogen ions moved into the IMS per glucose decomposed we can calculate the hypothetical number of ATP molecules produced per glucose molecule decomposed.
With new insight into the structure and function of the ETS complexes, plus the role of translocators, our calculations of the number of ATP molecules synthesized per glucose molecule has changed. Many students are still taught that 36-38 ATPs are formed per glucose. The calculations leading to these values are based on a an outdated understanding of the mechanisms involved in the ETS. We currently believe 30-32 is the range of ATPs produced as explained below.
Source of Hydrogen Ions
FADH2 in matrix
Two molecules of FADH2 are produced in the matrix during Krebs' cycle. These transfer their electrons to complex II where each generates a molecule of coenzyme QH2. Each reduced coenzyme transfers it's electrons to complex III and the electron transfer from that point on only adds six (6) hydrogen ions to the IMS. Because it requires four (4) hydrogen ions to generate one (1) ATP then each FADH2 contributes sufficient hydrogen ions to produce one and a half (6/4=1.5) ATP molecules.
NADH in matrix
Each NADH in the matrix will add 10 hydrogen ions to the IMS. Because it requires four (4) hydrogen ions to generate one (1) ATP then each NADH contributes sufficient hydrogen ions to produce two and a half (10/4=2.5) ATP molecules.
NADH in cytoplasm
This is where the variation in numbers of ATPs formed comes into play. Two NADH molecules are formed in the cytoplasm. Each must use a shuttle to deliver their electrons to the ETS. Which shuttle is used is up to chance.
- Use of the malate-aspartate shuttle delivers one NADH to the matrix where it adds ten (10) hydrogen ions to the IMS. AND/OR
- Use of the glycerol phosphate shuttle transfers a pair of electrons to coenzyme QH2. From here, electrons are transferred to complex III and the electron transfer from that point on only adds six (6) hydrogen ions to the IMS.
Final Calculations
Regardless of which shuttle is used by the two (2) cytoplasmic NADH molecules, there are eight (8) NADH molecules that will produce (8x2.5=20) twenty(20) ATP molecules. There are also two (2) FADH2 molecules that will produce (2x1.5=3) three (3) ATP molecules. This totals twenty-three (23).
- If both cytoplasmic NADH molecules use the more efficient malate-aspartate shuttle that produces one NADH then (2x2.5=5) five (5) more ATP molecules are added to the original twenty-three adding up to 28.
- If both cytoplasmic NADH molecules use the less efficient glycerol phosphate shuttle that produces one coenzyme QH2 then (2x1.5=3) three more ATP molecules are added to the original twenty-three adding up to (26).
Substrate-level Phosphorylation
There were two ATP molecules produced directly by during glycolysis. There were two more NTP (translate ATP) molecules produced during Krebs' cycle. Adding these four (4) to the 26-28 range calculated for ATP molecules produced by oxidative phosphorylation gives a final range of 30-32 per molecule of glucose broken down.