Electron Transport System & Oxidative Phosphorylation
The details of the molecular complexes involved in this set of reactions have recently been illuminated
in great detail. However, the mechanisms of the hydrogen ion 'pumps' associated with complexes I, II and IV
are still being investigated. Keep in mind that, at this point, the literature still contains
many versions of how this system works and the number of ATP molecules resulting per breakdown of one
glucose molecule. This summary will only consider the system as presented here.
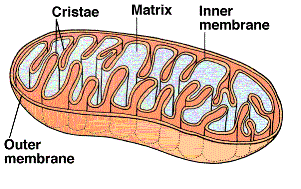
 The final phase occurs within the mitochondria. The cut-away illustration of a mitochondrion shows a large
inner membrane with folds called cristae. The center is a fluid-filled matrix that contains the soluble
enzymes of Krebs' cycle. There is a narrow space
between the inner and outer membranes (not labeled) called the intermembrane space (IMS).
The final phase occurs within the mitochondria. The cut-away illustration of a mitochondrion shows a large
inner membrane with folds called cristae. The center is a fluid-filled matrix that contains the soluble
enzymes of Krebs' cycle. There is a narrow space
between the inner and outer membranes (not labeled) called the intermembrane space (IMS).
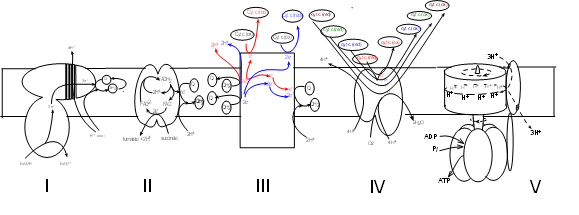
The schematic illustrations shows five large protein complexes that are embedded within
the inner mitochondrial membrane.
Current research suggests several ways they might be grouped but there is no consensus.
Many copies of each complex likely diffuse slowly within the membrane. Also within the inner
membrane are small hydrophobic molecules of coenzyme Q that diffuse more rapidly.
Water-soluble molecules of cytochrome c circulate within the intermembrane space (represented at the top
of the illustration). The mitochondrial matrix would be at the bottom of the illustration where the Roman
numerals are shown.
If considered as a set, the electron transport system (ETS) consists of four (I-IV) complexes plus
many molecules of coenzyme Q and cytochrome c. Complex V takes part in 'oxidative phosphorylation' -- the final step.
The details of this illustration are too small to read at
this resolution but each will be fully drawn out in the following tutorials.
Complex I
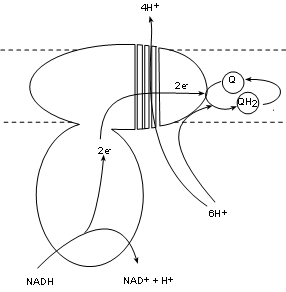 Complex I, also called NADH dehydrogenase, has a binding site for NADH that hangs into the matrix.
Two electrons are transferred into the complex while the hydrogen ion and the oxidized NAD+ are released into the matrix.
The electron pair is passed between several molecules within the complex to the complex surface buried
within the inner membrane. There is a bonding site there for coenzyme Q.
Complex I, also called NADH dehydrogenase, has a binding site for NADH that hangs into the matrix.
Two electrons are transferred into the complex while the hydrogen ion and the oxidized NAD+ are released into the matrix.
The electron pair is passed between several molecules within the complex to the complex surface buried
within the inner membrane. There is a bonding site there for coenzyme Q.
Existing within the hydrophobic interior of the inner membrane are small mobile molecules of coenzyme
Q. As common with electron transporters these have 'tag along' hydrogen nuclei (H+). These hydrogen
ions are picked
up directly from the matrix pool and join the electron pair to fully reduce coenzyme Q to QH2.
As this is occurring there is a conformational change -- possibly forming four channels -- that 'pumps' four (4) hydrogen ions from the
matrix pool into the intermembrane space.
The key points are:
One (1) NADH is oxidized and donates:
- one (1) hydrogen ion and NAD+ to the matrix.
- one (1) pair of electrons that the complex transfers to coenzyme Q forming Q-2 (not shown).
- Two (2) hydrogen ions from the matrix combine with Q-2 to form QH2.
- Four (4) hydrogen ions are 'pumped' from the matrix into the intermembrane space.
Complex II
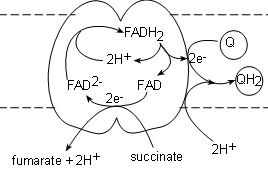 Complex II, also called succinate dehydrogenase , contains the actual enzyme we saw in step 6 of Krebs' cycle.
It's binding site for succinate is on the matrix side of the membrane and the transporter (FAD) is actually part
of the complex itself. The electrons from the succinate reduce FAD to FAD-2 while
the hydrogen ions and fumarate are released into the matrix.
Complex II, also called succinate dehydrogenase , contains the actual enzyme we saw in step 6 of Krebs' cycle.
It's binding site for succinate is on the matrix side of the membrane and the transporter (FAD) is actually part
of the complex itself. The electrons from the succinate reduce FAD to FAD-2 while
the hydrogen ions and fumarate are released into the matrix.
A FADH2, from within the complex, is oxidized producing a pair of hydrogen ions to complete
the reduction of the newly-formed FAD-2 thus regenerating itself. The FAD will accept the next pair of electrons
that are received. The electron pair is
transferred to a coenzyme Q attached to the surface of the complex to form Q-2. Finally, a pair of hydrogen
ions from the pool in the matrix completes the reduction of the coenzyme to QH2.
The key points are:
One succinate molecule is reduced to donate:
- One (1) pair of hydrogen ions and fumarate to the matrix.
- One (1) pair of electrons to the FAD within the complex that are eventually donated to
one (1) coenzyme Q forming Q-2 (not shown)
- One (1) pair of hydrogen ions from the matrix combines with the reduced Q-2 to form
one (1) QH2.
Complex III
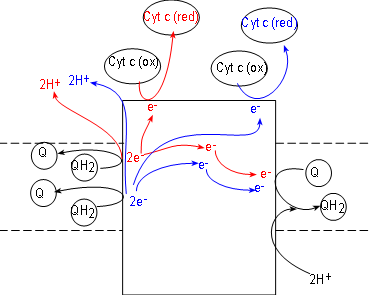 Initial reactants for this enzyme complex are coenzyme QH2 molecules (shown
at the left side of the complex). These have been accumulating in the hydrophobic interior of the inner
membrane due to the activities of complexes I and II.
Initial reactants for this enzyme complex are coenzyme QH2 molecules (shown
at the left side of the complex). These have been accumulating in the hydrophobic interior of the inner
membrane due to the activities of complexes I and II.
Complex III, also called coenzyme Q - cytochrome c reductase. simultaneously reduces both coenzyme Q
(illustrated at the right of the complex) and cytochrome c
(illustrated at the top of the complex). Cytochrome c molecules are numerous and located within the IMS.
Like complexes I and II, complex III also reduces coenzyme Q to QH2 (at the right of the
complex).
To understand what occurs we must follow one QH2
at a time. Inspect the top QH2 at the left of the complex. Notice it contacts the surface of the
complex to
release a pair of electrons (red) to the complex and the pair of hydrogen ions (red) into the
IMS.
Following the electrons (red arrows) notice that:
- One electron is transferred to the complex surface where it is picked up by an oxidized
cytochrome c (these receive only one electron). Being reduced (red), this cytochrome detaches from the
complex surface and returns to the IMS,
- The second electron (red) is transferred to the embedded part of the surface of the complex.
Here it will combine with a coenzyme Q attached there. This Q, with a single electron (not shown),
will remain attached to await the arrival of a second electron.
Now follow the products (blue) of the second QH2. A second pair of hydrogen ions
is added to the intermembrane space. Another electron (blue) reduces a second cytochrome c.
The second electron (blue) is transferred to the half-way reduced coenzyme Q plus a pair of hydrogen ions
from the matrix completes the formation of a molecule of QH2. It is released from the surface of complex III to
return to the hydrophobic interior of the membrane -- and complexes I and II.
The hydrogen ion 'pump' of this complex is indirect. In complexes I (and IV) hydrogen ions
are physically moved from the matrix to the space. In this case two pairs of hydrogen ions are moved out of
the interior of the inner membrane while one pair is moved from the matrix into the interior of that membrane.
*This is equivalent to moving a pair from the matrix to the space. Complex II is credited with pumping
one pair of hydrogen ions into the intermembrane space instead of two.
The key points are:
For each pair of QH2 oxidized::
- One (1) molecule of coenzyme Q is reduced.
- One (1) pair of hydrogen ions is removed from the matrix to help reduce Q-2.
- One (1) pair of hydrogen ions are moved from the matrix to the intermembrane space.*
- One (1) pair of cytochrome c transporters are reduced (one electron each).
Complex IV
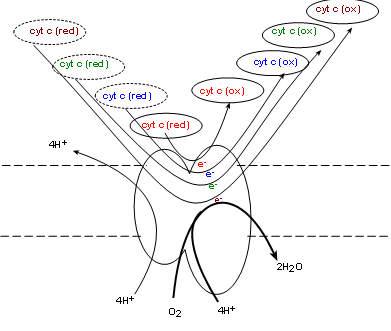 Complex IV, also called cytochrome c oxidase (COX) . will oxidize the reduced coenzyme c molecules produced
by complex III returning the oxidized forms to the intermembrane space -- and to complex III. Simultaneously, one molecule of
molecular oxygen (O2) from the matrix is taken into the complex. Current research indicates
four electrons, donated by four cytochrome c molecules, break the double-bond between the oxygen atoms
immediately forming two oxygen ions (O-2). The mechanism is unclear as to how four hydrogen
ions from the matrix are simultaneously
pumped into the intermembrane space. Each oxygen ion receives a pair of hydrogen ions from the matrix to
form water.
Oxygen is known as 'the final electron acceptor' reducing molecular oxygen to water.
Complex IV, also called cytochrome c oxidase (COX) . will oxidize the reduced coenzyme c molecules produced
by complex III returning the oxidized forms to the intermembrane space -- and to complex III. Simultaneously, one molecule of
molecular oxygen (O2) from the matrix is taken into the complex. Current research indicates
four electrons, donated by four cytochrome c molecules, break the double-bond between the oxygen atoms
immediately forming two oxygen ions (O-2). The mechanism is unclear as to how four hydrogen
ions from the matrix are simultaneously
pumped into the intermembrane space. Each oxygen ion receives a pair of hydrogen ions from the matrix to
form water.
Oxygen is known as 'the final electron acceptor' reducing molecular oxygen to water.
The key points are:
For every four (4) cytochrome c molecules that are oxidized the following occurs:
- Four (4) cytochrome c molecules are reduced
- Four (4) hydrogen ions are pumped from the matrix into the intermembrane space.
- One (1) molecular oxygen is reduced to water.
- Four (4) hydrogen ions are removed from the matrix to combine with oxygen.
Shuttles
There are two (2) NADH molecules produced in the cytoplasm during glycolysis. They cannot cross the inner membrane.
In order to utilize their potential to donate electrons they use one of two shuttle systems. These shuttles
are self replenishing so as not to run out of participating molecules.
Malate-Aspartate Shuttle
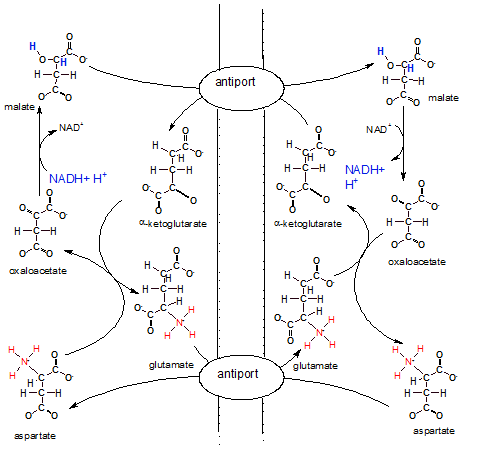 The illustration shows the inner membrane placed vertically with the IMS on it's left and the matrix on
it's right. There are two antiports (exchangers) embedded in the inner mitochondrial membrane that swap molecules
across the membrane.
The illustration shows the inner membrane placed vertically with the IMS on it's left and the matrix on
it's right. There are two antiports (exchangers) embedded in the inner mitochondrial membrane that swap molecules
across the membrane.
The overall big picture of all these reactions looks like two circles; a smaller one inside a larger one.
The arrows of the larger outer circle go clockwise while those of the inner one go counter-clockwise.
On the IMS side of the outer circle, aspartate is converted to oxaloacetate that is converted to
malate. On the matrix side the same conversions occur in the opposite direction.
On the IMS side of the small inner circle ,alpha-ketoglutarate is converted to glutamate while the opposite
conversions occur in the matrix.
Notice NADH + H+ in the IMS reducing oxaloacetate to malate. Malate now
is the bearer of the electrons and hydrogens with which we are concerned. To move these electrons and
hydrogens -- within malate --to the matrix there must be an alpha-ketoglutarate bound to the antiport molecule.
Only when both molecules --malate and alphaketoglutarate -- are attached will
the antiport simultaneously move them to opposite sides of the membrane. Once in the matrix the enzyme for step
8 of Krebs' cycle converts the malate to oxaloacetate releasing NADH and H+. Mission accomplished. Now
NADH can donate it's electrons to complex I of the ETS.
The matrix loses a molecule of alpha-ketoglutarate, an important substrate in Krebs' cycle, during the above
event. To compensate for this loss a new molecule is formed when oxaloacetate (formerly malate) reacts
with glutamate that has been transferred in from the IMS. During this reaction aspartate is formed. An antiport
for glutamate and aspartate insures a continuing supply of glutamate for future matrix reactions.
Inspection of the IMS side of the membrane shows that alpha-ketoglutarate is converted to glutamate to
insure it's continuing supply. Since it is moved to the matrix by an antiport there must also be a
continuing supply of its 'partner' aspartate. Aspartate is produced from oxaloacetate as mentioned earlier.
Also notice that once in the IMS the aspartate is converted to oxaloacetate to continue the outer-circle
reactions. A lot of work to move regenerate a single NADH on the matrix side of the membrane.
Glycerol Phosphate Shuttle
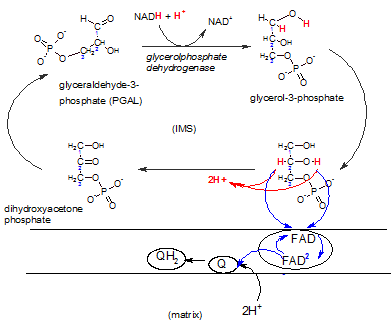 This shuttle system is primarily found in brown fat and infants.
A NADH produced in the cytoplasm during glycolysis can react with DHAP reducing it to
glycerol-3-phosphate that now carries the electrons and hydrogens.
The enzyme complex embedded in the inner membrane (lower right) contains the enzyme FAD-dependent
glyceolphosphate dehydrogenase. There is a binding site for glycerol-3-phosphate on it's IMS side.
The enzyme oxidizes this reactant while reducing FAD that is
an integral part of the complex. Notice that FAD accepts only the electron pair (blue) ; the hydrogen ion pair
(red) is set free in the IMS. The FAD-2 then reduces a coenzyme Q within the hydrophobic inner membrane
; this picks up a pair of hydrogen ions from the matrix to form
QH2 -- now available to react with complex III of the ETS.
This shuttle system is primarily found in brown fat and infants.
A NADH produced in the cytoplasm during glycolysis can react with DHAP reducing it to
glycerol-3-phosphate that now carries the electrons and hydrogens.
The enzyme complex embedded in the inner membrane (lower right) contains the enzyme FAD-dependent
glyceolphosphate dehydrogenase. There is a binding site for glycerol-3-phosphate on it's IMS side.
The enzyme oxidizes this reactant while reducing FAD that is
an integral part of the complex. Notice that FAD accepts only the electron pair (blue) ; the hydrogen ion pair
(red) is set free in the IMS. The FAD-2 then reduces a coenzyme Q within the hydrophobic inner membrane
; this picks up a pair of hydrogen ions from the matrix to form
QH2 -- now available to react with complex III of the ETS.
The difference between using this shuttle as opposed to the previous shuttle is that the previous one releases
the electron pair to complex I -- early in the ETS -- while this shuttle releases the electrons to complex III.
Shuttle Summary
The point of knowing which shuttle is used to capture the electron-transferring power of cytoplasmic
NADH molecules comes into play when trying to determine the number of ATP molecules formed as a result
of events occurring in the ETS. This will be discussed after the following section. The key point is that
if the malate-aspartate shuttle is used there is a NADH regenerated from a NADH. If the glycerol phosphate
shuttle is used a QH2 is produced. NAD enters the ETS at complex I so that it's
electron pair will pass through all three proton-pumping complexes. However, QH2 gives it's
electron pair to complex III bypassing the proton-pumping power of complex I.
Oxidative Phosphorylation
This group of reactions utilizes the hydrogen ion gradient across the inner membrane to produce ATP.
The term 'oxidative' refers to the fact that the hydrogen ions were concentrated on the IMS side of the
membrane due to the oxidation of the reduced transporters previously formed. 'Phosphorylation' refers to the
addition of a phosphate group to ADP forming ATP.
Complex V
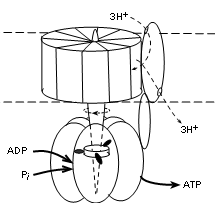 The illustration shows that this complex has two major parts; that embedded within the membrane (between
dotted lines) and that suspended into the matrix (lower part of illustration). The embedded part has a
central shaft that extends into the matrix part. This complex is actually a motor -- it moves!
The embedded part rotates -- staff included. The outer portion of the matrix part is stationary but the
shaft within it does move. The 'fuel' for the rotation is the diffusion of hydrogen ions, along their
electrochemical gradient (proton motive force), from the IMS (top of illustration) to the matrix.
The illustration shows that this complex has two major parts; that embedded within the membrane (between
dotted lines) and that suspended into the matrix (lower part of illustration). The embedded part has a
central shaft that extends into the matrix part. This complex is actually a motor -- it moves!
The embedded part rotates -- staff included. The outer portion of the matrix part is stationary but the
shaft within it does move. The 'fuel' for the rotation is the diffusion of hydrogen ions, along their
electrochemical gradient (proton motive force), from the IMS (top of illustration) to the matrix.
Current literature reports ten (10) proteins encircling the shaft within the membrane in human
mitochondria. When there are ten, research suggests that a set of three (3) hydrogen ions, diffusing through
a separate protein 'channel' will turn the membrane portion 120 degrees. It appears that
the number of proteins encircling this part of the shaft determines the number
of hydrogen ions required to produce this rotation.
There are three (3) sets of protein pairs (alpha and beta, not labeled) that fit side-by-side surrounding
the shaft in the matrix part. The rotation of the
shaft causes shape changes of the binding sites in these pairs.
The shaft between the three unit pairs has three 'spikes' arranged so that one contacts each pair
simultaneously (see illustration). With each
120 degree rotation the spikes rotate to contact the adjacent alpha/beta pair. Another 120 degree rotation moves them
to the next pair, etc. When spike #1 contacts a unit physical conformation of the pair changes to allow
an ADP and inorganic phosphate to enter. When spike #2 contacts this pair the bonding sites changes again
to bring the reactants into contact so ATP can be synthesized. Contact with spike
#3 separates the pair to release ATP and the return of spike #1 alters the conformation to accept
the next pair of reactants.
Each alpha/beta pair is 'one-step-behind' the one before it thus there is an ATP released with each
120 degree turn of the shaft. The stoichiometry (i.e., relation between the amount of two things) between
the diffusion of three (3) hydrogen ions is the production of one (1) ATP -- 3H+:1ATP
Translocators
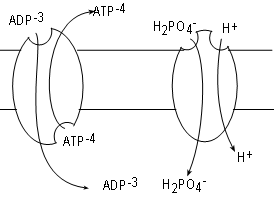 ADP is formed in the cytoplasm and has too great a negative charge to pass through the inner mitochondrial membrane. Likewise,
ATP formed within the matrix can not pass through either for the same reason. Many copies of a
transporter called
adenosine nucleotide translocase (ANT), embedded in the inner membrane, serve the exchange one molecule of ADP for
one molecule of ATP.
ADP is formed in the cytoplasm and has too great a negative charge to pass through the inner mitochondrial membrane. Likewise,
ATP formed within the matrix can not pass through either for the same reason. Many copies of a
transporter called
adenosine nucleotide translocase (ANT), embedded in the inner membrane, serve the exchange one molecule of ADP for
one molecule of ATP.
The mechanism for the exchange involves conformational changes. At any time a bonding site exists on
one side or the other but not both simultaneously as implied by the illustration. A site on the matrix side will bind ATP. Once bound the
molecule changes to evert the ATP to the IMS. After that a new site for ADP forms on the IMS side. When an
ADP binds the eversion takes place in reverse. Take note that the exchange creates a change in the charge (voltage)
difference across the membrane.
Inorganic phosphate is also in the cytoplasm but cannot pass through the membrane due to it's charge.
Multiple copies
of the translocator phosphate translocase, embedded in the membrane, relocate one inorganic phosphate plus
a hydrogen ion from the IMS into the matrix. In this case both substrates move in the same direction. This
is termed 'symport'; both substrates must be bound to activate the 'pump'. The driving force is the hydrogen
ion gradient favoring transport from the IMS to the matrix. Current literature favors counting the transfer
of this hydrogen ion into the matrix along with the three moving through complex V when tabulating the
number of ATPs formed.
ATP Synthesis
Synthesis of ATP required the movement of hydrogen ions along a gradient moving them from the IMS to
the matrix. This gradient is produced by the pumping of hydrogen ions into the IMS as described above.
We now believe it requires the movement of four (4) hydrogen ions -- three through complex V plus one
via the phosphate/hydrogen ion symport -- back into the matrix to synthesize one (1) ATP.
By tabulating the number of hydrogen ions moved into the IMS per glucose decomposed we can calculate the
hypothetical number of ATP molecules produced per glucose molecule decomposed.
With new insight into the structure and function of the ETS complexes, plus the role of translocators, our
calculations of the number of ATP molecules synthesized per glucose molecule has changed. Many students are
still taught that 36-38 ATPs are formed per glucose. The calculations leading to these values are
based on a an outdated understanding of the mechanisms involved in the ETS. We currently believe 30-32
is the range of ATPs produced as explained below.
Source of Hydrogen Ions
FADH2 in matrix
Two molecules of FADH2 are produced in the matrix during Krebs' cycle. These transfer their
electrons to complex II where each generates a molecule of coenzyme QH2. Each reduced coenzyme
transfers it's electrons to complex III and the electron transfer from that point on only adds six (6) hydrogen
ions to the IMS. Because it requires four (4) hydrogen ions to generate one (1) ATP then each FADH2
contributes sufficient hydrogen ions to produce one and a half (6/4=1.5) ATP molecules.
NADH in matrix
Each NADH in the matrix will add 10 hydrogen ions to the IMS. Because it requires four (4) hydrogen
ions to generate one (1) ATP then each NADH contributes sufficient hydrogen ions to produce two and a half
(10/4=2.5) ATP molecules.
NADH in cytoplasm
This is where the variation in numbers of ATPs formed comes into play.
Two NADH molecules are formed in the cytoplasm. Each must use a shuttle to deliver their electrons to the ETS.
Which shuttle is used is up to chance.
- Use of the malate-aspartate shuttle delivers one
NADH to the matrix where it adds ten (10) hydrogen ions to the IMS.
AND/OR
- Use of the glycerol phosphate shuttle transfers a pair
of electrons to coenzyme QH2. From here, electrons are transferred to complex III and the
electron transfer from that point on only adds six (6) hydrogen ions to the IMS.
Final Calculations
Regardless of which shuttle is used by the two (2) cytoplasmic NADH molecules, there are eight (8) NADH molecules
that will produce (8x2.5=20) twenty(20) ATP molecules. There are also two (2) FADH2 molecules
that will produce (2x1.5=3) three (3) ATP molecules. This totals twenty-three (23).
- If both cytoplasmic NADH molecules use the more efficient malate-aspartate shuttle that produces one NADH
then (2x2.5=5) five (5) more ATP molecules are added to the original twenty-three adding up to 28.
- If both cytoplasmic NADH molecules use the less efficient glycerol phosphate shuttle that produces one
coenzyme QH2 then (2x1.5=3) three more ATP molecules are added to the original twenty-three
adding up to (26).
Substrate-level Phosphorylation
There were two ATP molecules produced directly by during glycolysis. There
were two more NTP (translate ATP) molecules produced during Krebs' cycle. Adding
these four (4) to the 26-28 range calculated for ATP molecules produced by oxidative phosphorylation
gives a final range of 30-32 per molecule of glucose broken down.

 The final phase occurs within the mitochondria. The cut-away illustration of a mitochondrion shows a large
inner membrane with folds called cristae. The center is a fluid-filled matrix that contains the soluble
enzymes of Krebs' cycle. There is a narrow space
between the inner and outer membranes (not labeled) called the intermembrane space (IMS).
The final phase occurs within the mitochondria. The cut-away illustration of a mitochondrion shows a large
inner membrane with folds called cristae. The center is a fluid-filled matrix that contains the soluble
enzymes of Krebs' cycle. There is a narrow space
between the inner and outer membranes (not labeled) called the intermembrane space (IMS). Complex I, also called NADH dehydrogenase, has a binding site for NADH that hangs into the matrix.
Two electrons are transferred into the complex while the hydrogen ion and the oxidized NAD+ are released into the matrix.
The electron pair is passed between several molecules within the complex to the complex surface buried
within the inner membrane. There is a bonding site there for coenzyme Q.
Complex I, also called NADH dehydrogenase, has a binding site for NADH that hangs into the matrix.
Two electrons are transferred into the complex while the hydrogen ion and the oxidized NAD+ are released into the matrix.
The electron pair is passed between several molecules within the complex to the complex surface buried
within the inner membrane. There is a bonding site there for coenzyme Q.  Complex II, also called succinate dehydrogenase , contains the actual enzyme we saw in step 6 of Krebs' cycle.
It's binding site for succinate is on the matrix side of the membrane and the transporter (FAD) is actually part
of the complex itself. The electrons from the succinate reduce FAD to FAD-2 while
the hydrogen ions and fumarate are released into the matrix.
Complex II, also called succinate dehydrogenase , contains the actual enzyme we saw in step 6 of Krebs' cycle.
It's binding site for succinate is on the matrix side of the membrane and the transporter (FAD) is actually part
of the complex itself. The electrons from the succinate reduce FAD to FAD-2 while
the hydrogen ions and fumarate are released into the matrix.  Initial reactants for this enzyme complex are coenzyme QH2 molecules (shown
at the left side of the complex). These have been accumulating in the hydrophobic interior of the inner
membrane due to the activities of complexes I and II.
Initial reactants for this enzyme complex are coenzyme QH2 molecules (shown
at the left side of the complex). These have been accumulating in the hydrophobic interior of the inner
membrane due to the activities of complexes I and II. Complex IV, also called cytochrome c oxidase (COX) . will oxidize the reduced coenzyme c molecules produced
by complex III returning the oxidized forms to the intermembrane space -- and to complex III. Simultaneously, one molecule of
molecular oxygen (O2) from the matrix is taken into the complex. Current research indicates
four electrons, donated by four cytochrome c molecules, break the double-bond between the oxygen atoms
immediately forming two oxygen ions (O-2). The mechanism is unclear as to how four hydrogen
ions from the matrix are simultaneously
pumped into the intermembrane space. Each oxygen ion receives a pair of hydrogen ions from the matrix to
form water.
Oxygen is known as 'the final electron acceptor' reducing molecular oxygen to water.
Complex IV, also called cytochrome c oxidase (COX) . will oxidize the reduced coenzyme c molecules produced
by complex III returning the oxidized forms to the intermembrane space -- and to complex III. Simultaneously, one molecule of
molecular oxygen (O2) from the matrix is taken into the complex. Current research indicates
four electrons, donated by four cytochrome c molecules, break the double-bond between the oxygen atoms
immediately forming two oxygen ions (O-2). The mechanism is unclear as to how four hydrogen
ions from the matrix are simultaneously
pumped into the intermembrane space. Each oxygen ion receives a pair of hydrogen ions from the matrix to
form water.
Oxygen is known as 'the final electron acceptor' reducing molecular oxygen to water. The illustration shows the inner membrane placed vertically with the IMS on it's left and the matrix on
it's right. There are two antiports (exchangers) embedded in the inner mitochondrial membrane that swap molecules
across the membrane.
The illustration shows the inner membrane placed vertically with the IMS on it's left and the matrix on
it's right. There are two antiports (exchangers) embedded in the inner mitochondrial membrane that swap molecules
across the membrane. This shuttle system is primarily found in brown fat and infants.
A NADH produced in the cytoplasm during glycolysis can react with DHAP reducing it to
glycerol-3-phosphate that now carries the electrons and hydrogens.
The enzyme complex embedded in the inner membrane (lower right) contains the enzyme FAD-dependent
glyceolphosphate dehydrogenase. There is a binding site for glycerol-3-phosphate on it's IMS side.
The enzyme oxidizes this reactant while reducing FAD that is
an integral part of the complex. Notice that FAD accepts only the electron pair (blue) ; the hydrogen ion pair
(red) is set free in the IMS. The FAD-2 then reduces a coenzyme Q within the hydrophobic inner membrane
; this picks up a pair of hydrogen ions from the matrix to form
QH2 -- now available to react with complex III of the ETS.
This shuttle system is primarily found in brown fat and infants.
A NADH produced in the cytoplasm during glycolysis can react with DHAP reducing it to
glycerol-3-phosphate that now carries the electrons and hydrogens.
The enzyme complex embedded in the inner membrane (lower right) contains the enzyme FAD-dependent
glyceolphosphate dehydrogenase. There is a binding site for glycerol-3-phosphate on it's IMS side.
The enzyme oxidizes this reactant while reducing FAD that is
an integral part of the complex. Notice that FAD accepts only the electron pair (blue) ; the hydrogen ion pair
(red) is set free in the IMS. The FAD-2 then reduces a coenzyme Q within the hydrophobic inner membrane
; this picks up a pair of hydrogen ions from the matrix to form
QH2 -- now available to react with complex III of the ETS. The illustration shows that this complex has two major parts; that embedded within the membrane (between
dotted lines) and that suspended into the matrix (lower part of illustration). The embedded part has a
central shaft that extends into the matrix part. This complex is actually a motor -- it moves!
The embedded part rotates -- staff included. The outer portion of the matrix part is stationary but the
shaft within it does move. The 'fuel' for the rotation is the diffusion of hydrogen ions, along their
electrochemical gradient (proton motive force), from the IMS (top of illustration) to the matrix.
The illustration shows that this complex has two major parts; that embedded within the membrane (between
dotted lines) and that suspended into the matrix (lower part of illustration). The embedded part has a
central shaft that extends into the matrix part. This complex is actually a motor -- it moves!
The embedded part rotates -- staff included. The outer portion of the matrix part is stationary but the
shaft within it does move. The 'fuel' for the rotation is the diffusion of hydrogen ions, along their
electrochemical gradient (proton motive force), from the IMS (top of illustration) to the matrix. ADP is formed in the cytoplasm and has too great a negative charge to pass through the inner mitochondrial membrane. Likewise,
ATP formed within the matrix can not pass through either for the same reason. Many copies of a
transporter called
adenosine nucleotide translocase (ANT), embedded in the inner membrane, serve the exchange one molecule of ADP for
one molecule of ATP.
ADP is formed in the cytoplasm and has too great a negative charge to pass through the inner mitochondrial membrane. Likewise,
ATP formed within the matrix can not pass through either for the same reason. Many copies of a
transporter called
adenosine nucleotide translocase (ANT), embedded in the inner membrane, serve the exchange one molecule of ADP for
one molecule of ATP.